- Record: found
- Abstract: found
- Article: found
Genetic Dissection Reveals the Role of Ash1 Domains in Counteracting Polycomb Repression
Abstract
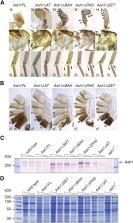
Antagonistic functions of Polycomb and Trithorax proteins are essential for proper development of all metazoans. While the Polycomb proteins maintain the repressed state of many key developmental genes, the Trithorax proteins ensure that these genes stay active in cells where they have to be expressed. Ash1 is the Trithorax protein that was proposed to counteract Polycomb repression by methylating lysine 36 of histone H3. However, it was recently shown that genetic replacement of Drosophila histone H3 with the variant that carried Arginine instead of Lysine at position 36 did not impair the ability of Ash1 to counteract Polycomb repression. This argues that Ash1 counteracts Polycomb repression by methylating yet unknown substrate(s) and that it is time to look beyond Ash1 methyltransferase SET domain, at other evolutionary conserved parts of the protein that received little attention. Here we used Drosophila genetics to demonstrate that Ash1 requires each of the BAH, PHD and SET domains to counteract Polycomb repression, while AT hooks are dispensable. Our findings argue that, in vivo, Ash1 acts as a multimer. Thereby it can combine the input of the SET domain and PHD-BAH cassette residing in different peptides. Finally, using new loss of function alleles, we show that zygotic Ash1 is required to prevent erroneous repression of homeotic genes of the bithorax complex in the embryo.
Most cited references43
- Record: found
- Abstract: found
- Article: not found
An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases.
- Record: found
- Abstract: found
- Article: not found
A gene complex controlling segmentation in Drosophila.
- Record: found
- Abstract: found
- Article: not found
