- Record: found
- Abstract: found
- Article: found
DNA barcoding identifies a cosmopolitan diet in the ocean sunfish

Read this article at
Abstract
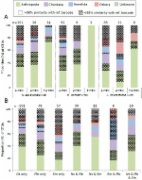
The ocean sunfish ( Mola mola) is the world’s heaviest bony fish reaching a body mass of up to 2.3 tonnes. However, the prey M. mola consumes to fuel this prodigious growth remains poorly known. Sunfish were thought to be obligate gelatinous plankton feeders, but recent studies suggest a more generalist diet. In this study, through molecular barcoding and for the first time, the diet of sunfish in the north-east Atlantic Ocean was characterised. Overall, DNA from the diet content of 57 individuals was successfully amplified, identifying 41 different prey items. Sunfish fed mainly on crustaceans and teleosts, with cnidarians comprising only 16% of the consumed prey. Although no adult fishes were sampled, we found evidence for an ontogenetic shift in the diet, with smaller individuals feeding mainly on small crustaceans and teleost fish, whereas the diet of larger fish included more cnidarian species. Our results confirm that smaller sunfish feed predominantly on benthic and on coastal pelagic species, whereas larger fish depend on pelagic prey. Therefore, sunfish is a generalist predator with a greater diversity of links in coastal food webs than previously realised. Its removal as fisheries’ bycatch may have wider reaching ecological consequences, potentially disrupting coastal trophic interactions.
Related collections
Most cited references44
- Record: found
- Abstract: not found
- Book Chapter: not found
The Effects of Fishing on Marine Ecosystems
- Record: found
- Abstract: found
- Article: not found