- Record: found
- Abstract: found
- Article: found
Cardiac arrhythmia and neuroexcitability gene variants in resected brain tissue from patients with sudden unexpected death in epilepsy (SUDEP)
Read this article at
Abstract
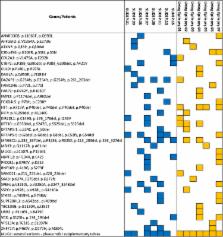
Sudden unexpected death in epilepsy (SUDEP) is the leading cause of epilepsy-related mortality in young adults. The exact mechanisms are unknown but death often follows a generalized tonic–clonic seizure. Proposed mechanisms include seizure-related respiratory, cardiac, autonomic, and arousal dysfunction. Genetic drivers underlying SUDEP risk are largely unknown. To identify potential SUDEP risk genes, we compared whole-exome sequences (WES) derived from formalin-fixed paraffin embedded surgical brain specimens of eight epilepsy patients who died from SUDEP with seven living controls matched for age at surgery, sex, year of surgery and lobe of resection. We compared identified variants from both groups filtering known polymorphisms from publicly available data as well as scanned for epilepsy and candidate SUDEP genes. In the SUDEP cohort, we identified mutually exclusive variants in genes involved in µ-opiod signaling, gamma-aminobutyric acid (GABA) and glutamate-mediated synaptic signaling, including ARRB2, ITPR1, GABRR2, SSTR5, GRIK1, CTNAP2, GRM8, GNAI2 and GRIK5. In SUDEP patients we also identified variants in genes associated with cardiac arrhythmia, including KCNMB1, KCNIP1, DPP6, JUP, F2, and TUBA3D, which were not present in living epilepsy controls. Our data shows that genomic analysis of brain tissue resected for seizure control can identify potential genetic biomarkers of SUDEP risk.
Epilepsy: Spotting genetic drivers of sudden death
Gene variants associated with abnormal heart rhythm and neuronal excitability may increase the risk of Sudden Unexpected Death in Epilepsy (SUDEP). SUDEP is the most common cause of death directly related to epilepsy, but little is known about the risk factors and mechanisms through which seizures can lead to death. Daniel Friedman, Orrin Devinsky and colleagues at New York University Langone Medical Center, US, compared whole-exome sequences from brain tissue belonging to eight epilepsy patients who died from SUDEP and seven matched living controls who had brain tissue removed for epilepsy treatment. In the SUDEP cases they identified 13 rare gene variants involved in cardiac arrhythmia and excitatory neurotransmission as potential genetic biomarkers of SUDEP risk. Further understanding the genetic contribution to epilepsy-related mortality will help develop effective preventive strategies.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines.
- Record: found
- Abstract: found
- Article: not found
Sudden unexpected death in epilepsy: epidemiology, mechanisms, and prevention.
- Record: found
- Abstract: found
- Article: not found