- Record: found
- Abstract: found
- Article: found
Control of Cell Identity by the Nuclear Receptor HNF4 in Organ Pathophysiology
Read this article at
Abstract
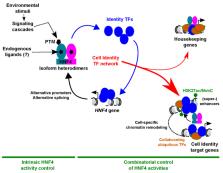
Hepatocyte Nuclear Factor 4 (HNF4) is a transcription factor (TF) belonging to the nuclear receptor family whose expression and activities are restricted to a limited number of organs including the liver and gastrointestinal tract. In this review, we present robust evidence pointing to HNF4 as a master regulator of cellular differentiation during development and a safekeeper of acquired cell identity in adult organs. Importantly, we discuss that transient loss of HNF4 may represent a protective mechanism upon acute organ injury, while prolonged impairment of HNF4 activities could contribute to organ dysfunction. In this context, we describe in detail mechanisms involved in the pathophysiological control of cell identity by HNF4, including how HNF4 works as part of cell-specific TF networks and how its expression/activities are disrupted in injured organs.
Related collections
Most cited references186
- Record: found
- Abstract: found
- Article: not found
Epithelial-mesenchymal transitions in development and disease.
- Record: found
- Abstract: found
- Article: not found