- Record: found
- Abstract: found
- Article: found
Multisite Phosphorylation Provides an Effective and Flexible Mechanism for Switch-Like Protein Degradation
Read this article at
Abstract
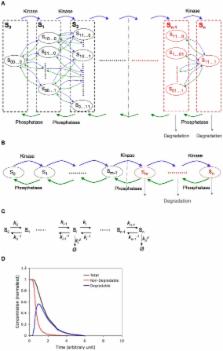
Phosphorylation-triggered degradation is a common strategy for elimination of regulatory proteins in many important cell signaling processes. Interesting examples include cyclin-dependent kinase inhibitors such as p27 in human and Sic1 in yeast, which play crucial roles during the G1/S transition in the cell cycle. In this work, we have modeled and analyzed the dynamics of multisite-phosphorylation-triggered protein degradation systematically. Inspired by experimental observations on the Sic1 protein and a previous intriguing theoretical conjecture, we develop a model to examine in detail the degradation dynamics of a protein featuring multiple phosphorylation sites and a threshold site number for elimination in response to a kinase signal. Our model explains the role of multiple phosphorylation sites, compared to a single site, in the regulation of protein degradation. A single-site protein cannot convert a graded input of kinase increase to much sharper output, whereas multisite phosphorylation is capable of generating a highly switch-like temporal profile of the substrate protein with two characteristics: a temporal threshold and rapid decrease beyond the threshold. We introduce a measure termed temporal response coefficient to quantify the extent to which a response in the time domain is switch-like and further investigate how this property is determined by various factors including the kinase input, the total number of sites, the threshold site number for elimination, the order of phosphorylation, the kinetic parameters, and site preference. Some interesting and experimentally verifiable predictions include that the non-degradable fraction of the substrate protein exhibits a more switch-like temporal profile; a sequential system is more switch-like, while a random system has the advantage of increased robustness; all the parameters, including the total number of sites, the threshold site number for elimination and the kinetic parameters synergistically determine the exact extent to which the degradation profile is switch-like. Our results suggest design principles for protein degradation switches which might be a widespread mechanism for precise regulation of cellular processes such as cell cycle progression.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
A methodology for performing global uncertainty and sensitivity analysis in systems biology.
- Record: found
- Abstract: found
- Article: not found
F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex.
- Record: found
- Abstract: found
- Article: not found