- Record: found
- Abstract: found
- Article: found
TP53 and MDM2 Gene Polymorphisms, Gene-Gene Interaction, and Hepatocellular Carcinoma Risk: Evidence from an Updated Meta-Analysis
Read this article at
Abstract
Background
The association between TP53 R72P and/or MDM2 SNP309 polymorphisms and hepatocellular carcinoma (HCC) risk has been widely reported, but results were inconsistent. To clarify the effects of these polymorphisms on HCC risk, an updated meta-analysis of all available studies was conducted.
Methods
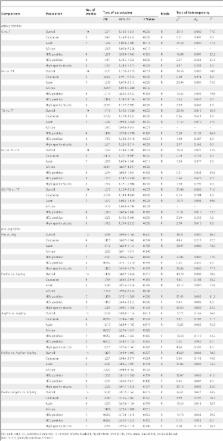
Eligible articles were identified by search of databases including PubMed, Cochrane Library, EMBASE and Chinese Biomedical Literature database (CBM) for the period up to July 2013. Data were extracted by two independent authors and pooled odds ratio (OR) with 95% confidence interval (CI) was calculated. Metaregression and subgroup analyses were performed to identify the source of heterogeneity.
Results
Finally, a total of 10 studies including 2,243 cases and 3,615 controls were available for MDM2 SNP309 polymorphism and 14 studies containing 4,855 cases and 6,630 controls were included for TP53 R72P polymorphism. With respect to MDM2 SNP309 polymorphism, significantly increased HCC risk was found in the overall population. In subgroup analysis by ethnicity and hepatitis virus infection status, significantly increased HCC risk was found in Asians, Caucasians, Africans, and HCV positive patients. With respect to TP53 R72P polymorphism, no significant association with HCC risk was observed in the overall and subgroup analyses. In the MDM2 SNP309–TP53 R72P interaction analysis, we found that subjects with MDM2 309TT and TP53 Pro/Pro genotype, MDM2 309 TG and TP53 Arg/Pro genotype, and MDM2 309 GG and TP53 Pro/Pro genotype were associated with significantly increased risk of developing HCC as compared with the reference MDM2 309TT and TP53 Arg/Arg genotype.
Conclusions
We concluded that MDM2 SNP309 polymorphism may play an important role in the carcinogenesis of HCC. In addition, our findings further suggest that the combination of MDM2 SNP 309 and TP53 Arg72Pro genotypes confers higher risk to develop HCC. Further large and well-designed studies are needed to confirm this association.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Suppression of induced pluripotent stem cell generation by the p53-p21 pathway.
- Record: found
- Abstract: found
- Article: not found
A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans.
- Record: found
- Abstract: found
- Article: not found