- Record: found
- Abstract: found
- Article: found
Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B
Read this article at
Abstract
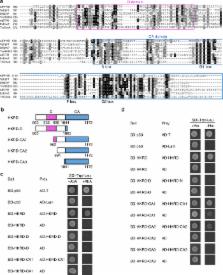
Plant phytochromes are thought to transduce light signals by mediating the degradation of phytochrome-interacting transcription factors (PIFs) through the N-terminal photosensory module, while the C-terminal module, including a histidine kinase-related domain (HKRD), does not participate in signaling. Here we show that the C-terminal module of Arabidopsis phytochrome B (PHYB) is sufficient to mediate the degradation of PIF3 specifically and to activate photosynthetic genes in the dark. The HKRD is a dimerization domain for PHYB homo and heterodimerization. A D1040V mutation, which disrupts the dimerization of HKRD and the interaction between C-terminal module and PIF3, abrogates PHYB nuclear accumulation, photobody biogenesis, and PIF3 degradation. By contrast, disrupting the interaction between PIF3 and PHYB’s N-terminal module has little effect on PIF3 degradation. Together, this study demonstrates that the dimeric form of the C-terminal module plays important signaling roles by targeting PHYB to subnuclear photobodies and interacting with PIF3 to trigger its degradation.
Abstract
Plant phytochromes mediate the degradation of PIF transcription factors to transduce light signaling. Here, contrary to previous models, Qiu et al. show that degradation of PIF3 does not require the N-terminal photosensory module of PHYB but can instead be mediated by the C-terminal output module.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
PIFs: pivotal components in a cellular signaling hub.
- Record: found
- Abstract: found
- Article: not found
Phytochrome structure and signaling mechanisms.
- Record: found
- Abstract: found
- Article: not found