- Record: found
- Abstract: found
- Article: found
SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals
Read this article at
Abstract
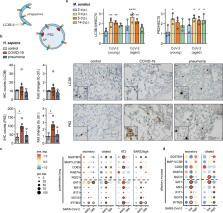
Viruses manipulate cellular metabolism and macromolecule recycling processes like autophagy. Dysregulated metabolism might lead to excessive inflammatory and autoimmune responses as observed in severe and long COVID-19 patients. Here we show that SARS-CoV-2 modulates cellular metabolism and reduces autophagy. Accordingly, compound-driven induction of autophagy limits SARS-CoV-2 propagation. In detail, SARS-CoV-2-infected cells show accumulation of key metabolites, activation of autophagy inhibitors (AKT1, SKP2) and reduction of proteins responsible for autophagy initiation (AMPK, TSC2, ULK1), membrane nucleation, and phagophore formation (BECN1, VPS34, ATG14), as well as autophagosome-lysosome fusion (BECN1, ATG14 oligomers). Consequently, phagophore-incorporated autophagy markers LC3B-II and P62 accumulate, which we confirm in a hamster model and lung samples of COVID-19 patients. Single-nucleus and single-cell sequencing of patient-derived lung and mucosal samples show differential transcriptional regulation of autophagy and immune genes depending on cell type, disease duration, and SARS-CoV-2 replication levels. Targeting of autophagic pathways by exogenous administration of the polyamines spermidine and spermine, the selective AKT1 inhibitor MK-2206, and the BECN1-stabilizing anthelmintic drug niclosamide inhibit SARS-CoV-2 propagation in vitro with IC 50 values of 136.7, 7.67, 0.11, and 0.13 μM, respectively. Autophagy-inducing compounds reduce SARS-CoV-2 propagation in primary human lung cells and intestinal organoids emphasizing their potential as treatment options against COVID-19.
Abstract
Viruses manipulate host cell pathways to support infection. Here the authors show that SARS-CoV-2 infection modulates cellular metabolism and limits autophagy, and identify druggable host pathways for virus inhibition.
Related collections
Most cited references83
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found