- Record: found
- Abstract: found
- Article: found
ATBF1 and NQO1 as candidate targets for allelic loss at chromosome arm 16q in breast cancer: Absence of somatic ATBF1 mutations and no role for the C609T NQO1 polymorphism
Read this article at
Abstract
Background
Loss of heterozygosity (LOH) at chromosome arm 16q is frequently observed in human breast cancer, suggesting that one or more target tumor suppressor genes (TSGs) are located there. However, detailed mapping of the smallest region of LOH has not yet resulted in the identification of a TSG at 16q. Therefore, the present study attempted to identify TSGs using an approach based on mRNA expression.
Methods
A cDNA microarray for the 16q region was constructed and analyzed using RNA samples from 39 breast tumors with known LOH status at 16q.
Results
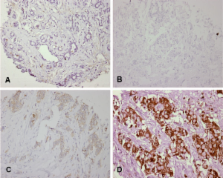
Five genes were identified to show lower expression in tumors with LOH at 16q compared to tumors without LOH. The genes for NAD(P)H dehydrogenase quinone ( NQO1) and AT-binding transcription factor 1 ( ATBF1) were further investigated given their functions as potential TSGs. NQO1 has been implicated in carcinogenesis due to its role in quinone detoxification and in stabilization of p53. One inactive polymorphic variant of NQO1 encodes a product showing reduced enzymatic activity. However, we did not find preferential targeting of the active NQO1 allele in tumors with LOH at 16q. Immunohistochemical analysis of 354 invasive breast tumors revealed that NQO1 protein expression in a subset of breast tumors is higher than in normal epithelium, which contradicts its proposed role as a tumor suppressor gene.
ATBF1 has been suggested as a target for LOH at 16q in prostate cancer. We analyzed the entire coding sequence in 48 breast tumors, but did not identify somatic sequence changes. We did find several in-frame insertions and deletions, two variants of which were reported to be somatic pathogenic mutations in prostate cancer. Here, we show that these variants are also present in the germline in 2.5% of 550 breast cancer patients and 2.9% of 175 healthy controls. This indicates that the frequency of these variants is not increased in breast cancer patients. Moreover, there is no preferential LOH of the wildtype allele in breast tumors.
Conclusion
Two likely candidate TSGs at 16q in breast cancer, NQO1 and ATBF1, were identified here as showing reduced expression in tumors with 16q LOH, but further analysis indicated that they are not target genes of LOH. Furthermore, our results call into question the validity of the previously reported pathogenic variants of the ATBF1 gene.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Mutation and cancer: statistical study of retinoblastoma.
- Record: found
- Abstract: found
- Article: not found