- Record: found
- Abstract: found
- Article: found
Persistence of frequently transmitted drug-resistant HIV-1 variants can be explained by high viral replication capacity
Read this article at
Abstract
Background
In approximately 10% of newly diagnosed individuals in Europe, HIV-1 variants harboring transmitted drug resistance mutations (TDRM) are detected. For some TDRM it has been shown that they revert to wild type while other mutations persist in the absence of therapy. To understand the mechanisms explaining persistence we investigated the in vivo evolution of frequently transmitted HIV-1 variants and their impact on in vitro replicative capacity.
Results
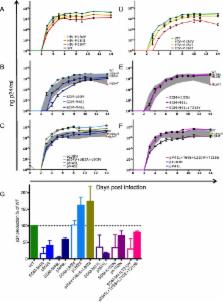
We selected 31 individuals infected with HIV-1 harboring frequently observed TDRM such as M41L or K103N in reverse transcriptase (RT) or M46L in protease. In all these samples, polymorphisms at non-TDRM positions were present at baseline (median protease: 5, RT: 6). Extensive analysis of viral evolution of protease and RT demonstrated that the majority of TDRM (51/55) persisted for at least a year and even up to eight years in the plasma. During follow-up only limited selection of additional polymorphisms was observed (median: 1).
To investigate the impact of frequently observed TDRM on the replication capacity, mutant viruses were constructed with the most frequently encountered TDRM as site-directed mutants in the genetic background of the lab strain HXB2. In addition, viruses containing patient-derived protease or RT harboring similar TDRM were made. The replicative capacity of all viral variants was determined by infecting peripheral blood mononuclear cells and subsequently monitoring virus replication. The majority of site-directed mutations (M46I/M46L in protease and M41L, M41L + T215Y and K103N in RT) decreased viral replicative capacity; only protease mutation L90M did not hamper viral replication. Interestingly, most patient-derived viruses had a higher in vitro replicative capacity than the corresponding site-directed mutant viruses.
Conclusions
We demonstrate limited in vivo evolution of protease and RT harbouring frequently observed TDRM in the plasma. This is in line with the high in vitro replication capacity of patient-derived viruses harbouring TDRM compared to site-directed mutant viruses harbouring TDRM. As site-directed mutant viruses have a lower replication capacity than the patient-derived viruses with similar mutational patterns, we propose that (baseline) polymorphisms function as compensatory mutations improving viral replication capacity.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006.
- Record: found
- Abstract: not found
- Article: not found
Update of the drug resistance mutations in HIV-1: March 2013.
- Record: found
- Abstract: found
- Article: not found