- Record: found
- Abstract: found
- Article: found
HIF3A association with adiposity: the story begins before birth
Read this article at
Abstract
Aim:
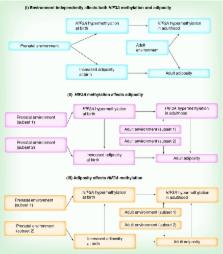
Determine if the association of HIF3A DNA methylation with weight and adiposity is detectable early in life.
Material & methods:
We determined HIF3A genotype and DNA methylation patterns (on hybridization arrays) in DNA extracted from umbilical cords of 991 infants. Methylation levels at three CpGs in the HIF3A first intron were related to neonatal and infant anthropometry and to genotype at nearby polymorphic sites.
Results & conclusion:
Higher methylation levels at three previously described HIF3A CpGs were associated with greater infant weight and adiposity. The effect sizes were slightly smaller than those reported for adult BMI. There was also an interaction within cis-genotype. The association between higher DNA methylation at HIF3A and increased adiposity is present in neonates. In this study, no particular prenatal factor strongly influenced HIF3A hypermethylation. Our data nonetheless suggest shared prenatal influences on HIF3A methylation and adiposity.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
High density DNA methylation array with single CpG site resolution.
- Record: found
- Abstract: found
- Article: not found
Epigenetic mechanisms that underpin metabolic and cardiovascular diseases.
- Record: found
- Abstract: not found
- Article: not found