- Record: found
- Abstract: found
- Article: not found
Hydrogen peroxide sensing, signaling and regulation of transcription factors

Read this article at
Abstract
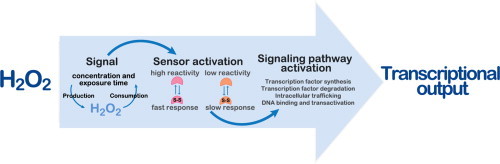
The regulatory mechanisms by which hydrogen peroxide (H 2O 2) modulates the activity of transcription factors in bacteria (OxyR and PerR), lower eukaryotes (Yap1, Maf1, Hsf1 and Msn2/4) and mammalian cells (AP-1, NRF2, CREB, HSF1, HIF-1, TP53, NF-κB, NOTCH, SP1 and SCREB-1) are reviewed. The complexity of regulatory networks increases throughout the phylogenetic tree, reaching a high level of complexity in mammalians. Multiple H 2O 2 sensors and pathways are triggered converging in the regulation of transcription factors at several levels: (1) synthesis of the transcription factor by upregulating transcription or increasing both mRNA stability and translation; (ii) stability of the transcription factor by decreasing its association with the ubiquitin E3 ligase complex or by inhibiting this complex; (iii) cytoplasm–nuclear traffic by exposing/masking nuclear localization signals, or by releasing the transcription factor from partners or from membrane anchors; and (iv) DNA binding and nuclear transactivation by modulating transcription factor affinity towards DNA, co-activators or repressors, and by targeting specific regions of chromatin to activate individual genes. We also discuss how H 2O 2 biological specificity results from diverse thiol protein sensors, with different reactivity of their sulfhydryl groups towards H 2O 2, being activated by different concentrations and times of exposure to H 2O 2. The specific regulation of local H 2O 2 concentrations is also crucial and results from H 2O 2 localized production and removal controlled by signals. Finally, we formulate equations to extract from typical experiments quantitative data concerning H 2O 2 reactivity with sensor molecules. Rate constants of 140 M −1 s −1 and ≥1.3 × 10 3 M −1 s −1 were estimated, respectively, for the reaction of H 2O 2 with KEAP1 and with an unknown target that mediates NRF2 protein synthesis. In conclusion, the multitude of H 2O 2 targets and mechanisms provides an opportunity for highly specific effects on gene regulation that depend on the cell type and on signals received from the cellular microenvironment.
Highlights
-
•
Complexity of redox regulation increases along the phylogenetic tree.
-
•
Complex regulatory networks allow for a high degree of H 2O 2 biological plasticity.
-
•
H 2O 2 modulates gene expression at all steps from transcription to protein synthesis.
-
•
Fast response (s) is mediated by sensors with high H 2O 2 reactivity.
-
•
Low reactivity H 2O 2 sensors may mediate slow (h) or localized H 2O 2 responses.
Graphical Abstract
Related collections
Most cited references331
- Record: found
- Abstract: found
- Article: not found
The canonical Notch signaling pathway: unfolding the activation mechanism.
- Record: found
- Abstract: found
- Article: not found
Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B.
- Record: found
- Abstract: found
- Article: not found