- Record: found
- Abstract: found
- Article: found
Sequence analysis of Plasmodium vivax Duffy binding proteins reveals the presence of unique haplotypes and diversifying selection in Ethiopian isolates
Read this article at
Abstract
Background
Red blood cell invasion by the Plasmodium vivax merozoite requires interaction between the Duffy antigen receptor for chemokines (DARC) and the P. vivax Duffy-binding protein II (PvDBPII). Given that the disruption of this interaction prevents P. vivax blood-stage infection, a PvDBP-based vaccine development has been well recognized. However, the polymorphic nature of PvDBPII prevents a strain transcending immune response and complicates attempts to design a vaccine.
Methods
Twenty-three P. vivax clinical isolates collected from three areas of Ethiopia were sequenced at the pvdbpII locus. A total of 392 global pvdbpII sequences from seven P. vivax endemic countries were also retrieved from the NCBI archive for comparative analysis of genetic diversity, departure from neutrality, linkage disequilibrium, genetic differentiation, PvDBP polymorphisms, recombination and population structure of the parasite population. To establish a haplotype relationship a network was constructed using the median joining algorithm.
Results
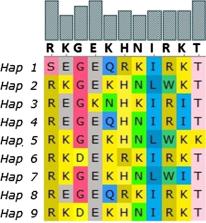
A total of 110 variable sites were found, of which 44 were parsimony informative. For Ethiopian isolates there were 12 variable sites of which 10 were parsimony informative. These parsimony informative variants resulted in 10 nonsynonymous mutations. The overall haplotype diversity for global isolates was 0.9596; however, the haplotype diversity was 0.874 for Ethiopia. Fst values for genetic revealed Ethiopian isolates were closest to Indian isolates as well as to Sri Lankan and Sudanese isolates but further away from Mexican, Papua New Guinean and South Korean isolates. There was a total of 136 haplotypes from the 415 global isolates included for this study. Haplotype prevalence ranged from 36.76% to 0.7%, from this 74.2% were represented by single parasite isolates. None of the Ethiopian isolates grouped with the Sal I reference haplotype. From the total observed nonsynonymous mutations 13 mapped to experimentally verified epitope sequences. Including 10 non-synonymous mutations from Ethiopia. However, all the polymorphic regions in Ethiopian isolates were located away from DARC, responsible for junction formation.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy.
- Record: found
- Abstract: found
- Article: not found
Evidence and implications of mortality associated with acute Plasmodium vivax malaria.

- Record: found
- Abstract: found
- Article: found