- Record: found
- Abstract: found
- Article: found
Cisplatin +/− rucaparib after preoperative chemotherapy in patients with triple-negative or BRCA mutated breast cancer
Read this article at
Abstract
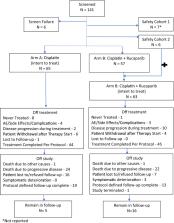
Patients with triple-negative breast cancer (TNBC) who have residual disease after neoadjuvant therapy have a high risk of recurrence. We tested the impact of DNA-damaging chemotherapy alone or with PARP inhibition in this high-risk population. Patients with TNBC or deleterious BRCA mutation (TNBC/BRCAmut) who had >2 cm of invasive disease in the breast or persistent lymph node (LN) involvement after neoadjuvant therapy were assigned 1:1 to cisplatin alone or with rucaparib. Germline mutations were identified with BROCA analysis. The primary endpoint was 2-year disease-free survival (DFS) with 80% power to detect an HR 0.5. From Feb 2010 to May 2013, 128 patients were enrolled. Median tumor size at surgery was 1.9 cm (0–11.5 cm) with 1 (0–38) involved LN; median Residual Cancer Burden (RCB) score was 2.6. Six patients had known deleterious BRCA1 or BRCA2 mutations at study entry, but BROCA identified deleterious mutations in 22% of patients with available samples. Toxicity was similar in both arms. Despite frequent dose reductions (21% of patients) and delays (43.8% of patients), 73% of patients completed planned cisplatin. Rucaparib exposure was limited with median concentration 275 (82–4694) ng/mL post-infusion on day 3. The addition of rucaparib to cisplatin did not increase 2-year DFS (54.2% cisplatin vs. 64.1% cisplatin + rucaparib; P = 0.29). In the high-risk post preoperative TNBC/BRCAmut setting, the addition of low-dose rucaparib did not improve 2-year DFS or increase the toxicity of cisplatin. Genetic testing was underutilized in this high-risk population.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis.
- Record: found
- Abstract: found
- Article: not found
Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies.
- Record: found
- Abstract: found
- Article: not found