- Record: found
- Abstract: found
- Article: found
Architecture of the RNA polymerase II–TFIIF complex revealed by cross-linking and mass spectrometry
Read this article at
Abstract
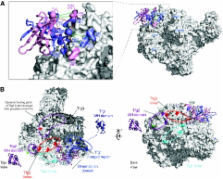
Higher-order multi-protein complexes such as RNA polymerase II (Pol II) complexes with transcription initiation factors are often not amenable to X-ray structure determination. Here, we show that protein cross-linking coupled to mass spectrometry (MS) has now sufficiently advanced as a tool to extend the Pol II structure to a 15-subunit, 670 kDa complex of Pol II with the initiation factor TFIIF at peptide resolution. The N-terminal regions of TFIIF subunits Tfg1 and Tfg2 form a dimerization domain that binds the Pol II lobe on the Rpb2 side of the active centre cleft near downstream DNA. The C-terminal winged helix (WH) domains of Tfg1 and Tfg2 are mobile, but the Tfg2 WH domain can reside at the Pol II protrusion near the predicted path of upstream DNA in the initiation complex. The linkers between the dimerization domain and the WH domains in Tfg1 and Tfg2 are located to the jaws and protrusion, respectively. The results suggest how TFIIF suppresses non-specific DNA binding and how it helps to recruit promoter DNA and to set the transcription start site. This work establishes cross-linking/MS as an integrated structure analysis tool for large multi-protein complexes.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites.
- Record: found
- Abstract: found
- Article: not found
Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
- Record: found
- Abstract: found
- Article: not found