- Record: found
- Abstract: found
- Article: found
Automatic Segmentation of Anatomical Structures from CT Scans of Thorax for RTP
Read this article at
Abstract
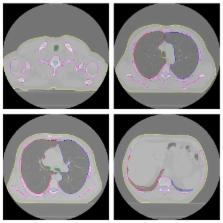
Modern radiotherapy techniques are vulnerable to delineation inaccuracies owing to the steep dose gradient around the target. In this aspect, accurate contouring comprises an indispensable part of optimal radiation treatment planning (RTP). We suggest a fully automated method to segment the lungs, trachea/main bronchi, and spinal canal accurately from computed tomography (CT) scans of patients with lung cancer to use for RTP. For this purpose, we developed a new algorithm for inclusion of excluded pathological areas into the segmented lungs and a modified version of the fuzzy segmentation by morphological reconstruction for spinal canal segmentation and implemented some image processing algorithms along with them. To assess the accuracy, we performed two comparisons between the automatically obtained results and the results obtained manually by an expert. The average volume overlap ratio values range between 94.30 ± 3.93% and 99.11 ± 0.26% on the two different datasets. We obtained the average symmetric surface distance values between the ranges of 0.28 ± 0.21 mm and 0.89 ± 0.32 mm by using the same datasets. Our method provides favorable results in the segmentation of CT scans of patients with lung cancer and can avoid heavy computational load and might offer expedited segmentation that can be used in RTP.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Comparison and evaluation of methods for liver segmentation from CT datasets.
- Record: found
- Abstract: found
- Article: not found
Active shape model segmentation with optimal features.
- Record: found
- Abstract: found
- Article: not found