- Record: found
- Abstract: found
- Article: found
Non-Coding RNAs and their Integrated Networks
Read this article at
Abstract
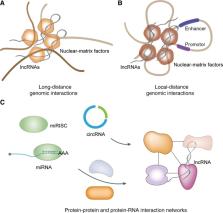
Eukaryotic genomes are pervasively transcribed. Besides protein-coding RNAs, there are different types of non-coding RNAs that modulate complex molecular and cellular processes. RNA sequencing technologies and bioinformatics methods greatly promoted the study of ncRNAs, which revealed ncRNAs’ essential roles in diverse aspects of biological functions. As important key players in gene regulatory networks, ncRNAs work with other biomolecules, including coding and non-coding RNAs, DNAs and proteins. In this review, we discuss the distinct types of ncRNAs, including housekeeping ncRNAs and regulatory ncRNAs, their versatile functions and interactions, transcription, translation, and modification. Moreover, we summarize the integrated networks of ncRNA interactions, providing a comprehensive landscape of ncRNAs regulatory roles.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
The transcriptional landscape of the mammalian genome.
- Record: found
- Abstract: found
- Article: not found
A coding-independent function of gene and pseudogene mRNAs regulates tumour biology
- Record: found
- Abstract: found
- Article: not found