- Record: found
- Abstract: found
- Article: found
Analysis of chromosomal abnormalities by CGH-array in patients with dysmorphic and intellectual disability with normal karyotype
research-article
Rodrigo Pratte-Santos
1
,
2 ,
Katyanne Heringer Ribeiro
2 ,
Thainá Altoe Santos
2 ,
Terezinha Sarquis Cintra
3
Jan-Mar 2016
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
ABSTRACT
Objective
To investigate chromosomal abnormalities by CGH-array in patients with dysmorphic features and intellectual disability with normal conventional karyotype.
Methods
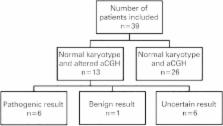
Retrospective study, carried out from January 2012 to February 2014, analyzing the CGH-array results of 39 patients.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
A high-resolution survey of deletion polymorphism in the human genome.
Donald F Conrad, T. Andrews, Nigel P Carter … (2006)
- Record: found
- Abstract: found
- Article: not found
Array comparative genomic hybridization and its applications in cancer.
Donna Albertson, Daniel Pinkel (2005)
- Record: found
- Abstract: found
- Article: not found