- Record: found
- Abstract: found
- Article: found
Molecular epidemiology of hepatitis B virus infection in Norway
Read this article at
Abstract
Background
Hepatitis B virus (HBV) infection remains a serious global health challenge. The widespread distribution of HBV is highlighted by multiple HBV genotypes associated with different geographical origin and transmission patterns, as well as, clinical outcomes. Investigating population HBV genotype composition and origin is therefore highly warranted.
Methods
In this molecular epidemiological study we analysed 1157 HBV S-gene sequences collected from patients in Norway, primarily in the period 2004–2011, and linked them to epidemiological data from the Norwegian surveillance system for communicable diseases.
Results
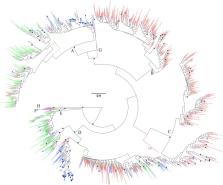
Of the patients with reported country of infection ( n = 909), 10% ( n = 93) were infected in Norway, but the majority ( n = 816; 90%) stated that they became infected outside of Norway. Of the patients infected outside of Norway, most became infected in Southeast and East Asia ( n = 465; 51%) and Central, West, and North Africa ( n = 254; 28%). The distribution of HBV genotypes in Norway is dominated by genotype D (32%) followed by genotype A (22%), B and C (18 and 18%, respectively), and E (7%). Genotype B, C and E were phylogenetically categorized by a majority of sequences originating from distinct geographical regions, either Asia or Africa, whereas genotype A and D originated from multiple geographic regions. However, within genotype A and D, our molecular epidemiology analysis indicated a geographical clustering of sequences depending on their geographical origin.
Conclusions
The majority of HBV patients in Norway became infected outside of Norway and were represented by most common genotypes. Patients stated to have been infected in Norway were found primarily within genotype A and D, and were phylogenetically characterized by both small local clusters and interspersed sequences that clustered with non-Norwegian sequences, indicating that a proportion of the patients assumed to have been infected in Norway likely became infected outside of Norway although assumed the contrary.
Related collections
Most cited references15
- Record: found
- Abstract: found
- Article: not found
Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes.
- Record: found
- Abstract: found
- Article: not found
Hepatitis B virus taxonomy and hepatitis B virus genotypes.
- Record: found
- Abstract: found
- Article: not found