- Record: found
- Abstract: found
- Article: found
Brainstem pathology in amyotrophic lateral sclerosis and primary lateral sclerosis: A longitudinal neuroimaging study
Read this article at
Highlights
-
•
Computational neuroimaging captures focal brainstem pathology in motor neuron diseases in contrast to both healthy- and disease controls.
-
•
ALS patients exhibit progressive medulla oblongata, pontine and mesencephalic volume loss over time.
-
•
Brainstem atrophy in ALS and PLS is dominated by medulla oblongata volume reductions.
-
•
Vertex analyses of ALS patients reveal flattening of the medullary pyramids bilaterally.
-
•
Morphometric analyses in ALS detect density reductions in the mesencephalic crura consistent with corticospinal tract degeneration.
Abstract
Background
Brainstem pathology is a hallmark feature of ALS, yet most imaging studies focus on cortical grey matter alterations and internal capsule white matter pathology. Brainstem imaging in ALS provides a unique opportunity to appraise descending motor tract degeneration and bulbar lower motor neuron involvement.
Methods
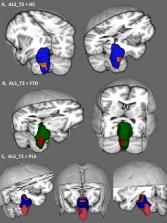
A prospective longitudinal imaging study has been undertaken with 100 patients with ALS, 33 patients with PLS, 30 patients with FTD and 100 healthy controls. Volumetric, vertex and morphometric analyses were conducted correcting for demographic factors to characterise disease-specific patterns of brainstem pathology. Using a Bayesian segmentation algorithm, the brainstem was segmented into the medulla, pons and mesencephalon to measure regional volume reductions, shape analyses were performed to ascertain the atrophy profile of each study group and region-of-interest morphometry was used to evaluate focal density alterations.
Results
ALS and PLS patients exhibit considerable brainstem atrophy compared to both disease- and healthy controls. Volume reductions in ALS and PLS are dominated by medulla oblongata pathology, but pontine atrophy can also be detected. In ALS, vertex analyses confirm the flattening of the medullary pyramids bilaterally in comparison to healthy controls and widespread pontine shape deformations in contrast to PLS. The ALS cohort exhibit bilateral density reductions in the mesencephalic crura in contrast to healthy controls, central pontine atrophy compared to disease controls, peri-aqueduct mesencephalic and posterior pontine changes in comparison to PLS patients.
Conclus
ions: Computational brainstem imaging captures the degeneration of both white and grey matter components in ALS. Our longitudinal data indicate progressive brainstem atrophy over time, underlining the biomarker potential of quantitative brainstem measures in ALS. At a time when a multitude of clinical trials are underway worldwide, there is an unprecedented need for accurate biomarkers to monitor disease progression and detect response to therapy. Brainstem imaging is a promising addition to candidate biomarkers of ALS and PLS.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis.
- Record: found
- Abstract: found
- Article: not found
Bayesian segmentation of brainstem structures in MRI.
- Record: found
- Abstract: found
- Article: not found