- Record: found
- Abstract: found
- Article: found
Anti-staphylococcal activities of lysostaphin and LytM catalytic domain
Read this article at
Abstract
Background
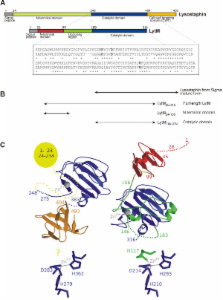
Lysostaphin and the catalytic domain of LytM cleave pentaglycine crossbridges of Staphylococcus aureus peptidoglycan. The bacteriocin lysostaphin is secreted by Staphylococcus simulans biovar staphylolyticus and directed against the cell walls of competing S. aureus. LytM is produced by S. aureus as a latent autolysin and can be activated in vitro by the removal of an N-terminal domain and occluding region.
Results
We compared the efficacies of the lysostaphin and LytM catalytic domains using a newly developed model of chronic S. aureus infected eczema. Lysostaphin was effective, like in other models. In contrast, LytM was not significantly better than control. The different treatment outcomes could be correlated with in vitro properties of the proteins, including proteolytic stability, affinity to cell wall components other than peptidoglycan, and sensitivity to the ionic milieu.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4.
- Record: found
- Abstract: found
- Article: not found
MEROPS: the peptidase database
- Record: found
- Abstract: found
- Article: not found