- Record: found
- Abstract: found
- Article: found
Rickettsia association with two Macrolophus (Heteroptera: Miridae) species: A comparative study of phylogenies and within-host localization patterns
Read this article at
Abstract
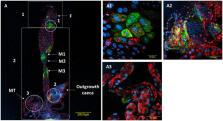
Many arthropods host bacterial symbionts, some of which are known to influence host nutrition and diet breadth. Omnivorous bugs of the genus Macrolophus (Heteroptera: Miridae) are mainly predatory, but may also feed on plants. The species M. pygmaeus and M. melanotoma (= M. caliginosus) are key natural enemies of various economically important agricultural pests, and are known to harbor two Rickettsia species, R. bellii and R. limoniae. To test for possible involvement of symbiotic bacteria in the nutritional ecology of these biocontrol agents, the abundance, phylogeny, and distribution patterns of the two Rickettsia species in M. pygmaeus and M. melanotoma were studied. Both of the Rickettsia species were found in 100 and 84% of all tested individuals of M. pygmaeus and M. melanotoma, respectively. Phylogenetic analysis showed that a co-evolutionary process between Macrolophus species and their Rickettsia is infrequent. Localization of R. bellii and R. limoniae has been detected in both female and male of M. pygmaeus and M. melanotoma. FISH analysis of female gonads revealed the presence of both Rickettsia species in the germarium of both bug species. Each of the two Rickettsia species displayed a unique distribution pattern along the digestive system of the bugs, mostly occupying separate epithelial cells, unknown caeca-like organs, the Malpighian tubules and the salivary glands. This pattern differed between the two Macrolophus species: in M. pygmaeus, R. limoniae was distributed more broadly along the host digestive system and R. bellii was located primarily in the foregut and midgut. In contrast, in M. melanotoma, R. bellii was more broadly distributed along the digestive system than the clustered R. limoniae. Taken together, these results suggest that Rickettsia may have a role in the nutritional ecology of their plant-and prey-consuming hosts.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.
- Record: found
- Abstract: found
- Article: not found
16S ribosomal DNA amplification for phylogenetic study.
- Record: found
- Abstract: not found
- Article: not found