- Record: found
- Abstract: found
- Article: found
IFITM Proteins Inhibit Entry Driven by the MERS-Coronavirus Spike Protein: Evidence for Cholesterol-Independent Mechanisms
Abstract
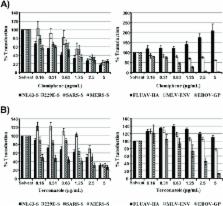
The interferon-inducible transmembrane (IFITM) proteins 1, 2 and 3 inhibit the host cell entry of several enveloped viruses, potentially by promoting the accumulation of cholesterol in endosomal compartments. IFITM3 is essential for control of influenza virus infection in mice and humans. In contrast, the role of IFITM proteins in coronavirus infection is less well defined. Employing a retroviral vector system for analysis of coronavirus entry, we investigated the susceptibility of human-adapted and emerging coronaviruses to inhibition by IFITM proteins. We found that entry of the recently emerged Middle East respiratory syndrome coronavirus (MERS-CoV) is sensitive to inhibition by IFITM proteins. In 293T cells, IFITM-mediated inhibition of cellular entry of the emerging MERS- and SARS-CoV was less efficient than blockade of entry of the globally circulating human coronaviruses 229E and NL63. Similar differences were not observed in A549 cells, suggesting that cellular context and/or IFITM expression levels can impact inhibition efficiency. The differential IFITM-sensitivity of coronaviruses observed in 293T cells afforded the opportunity to investigate whether efficiency of entry inhibition by IFITMs and endosomal cholesterol accumulation correlate. No such correlation was observed. Furthermore, entry mediated by the influenza virus hemagglutinin was robustly inhibited by IFITM3 but was insensitive to accumulation of endosomal cholesterol, indicating that modulation of cholesterol synthesis/transport did not account for the antiviral activity of IFITM3. Collectively, these results show that the emerging MERS-CoV is a target of the antiviral activity of IFITM proteins and demonstrate that mechanisms other than accumulation of endosomal cholesterol can contribute to viral entry inhibition by IFITMs.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
A decade after SARS: strategies for controlling emerging coronaviruses
- Record: found
- Abstract: found
- Article: not found
Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product.
- Record: found
- Abstract: found
- Article: not found