- Record: found
- Abstract: found
- Article: not found
Microbial pathway for anaerobic 5′-methylthioadenosine metabolism coupled to ethylene formation
Read this article at
Significance
Sulfur is an essential element required by all organisms. Therefore, salvage of wasteful, sulfur-containing cellular by-products can be critical. Methionine salvage pathways for organisms living in oxic environments are well established. However, if and by what mechanisms organisms living in anoxic environments can regenerate methionine from such by-products remain largely unknown. This work identifies a strictly anaerobic methionine salvage pathway, the key genes for which appear to be widespread among obligate and facultatively anaerobic bacteria. Strikingly, this pathway also results in the formation of ethylene gas, a key plant hormone and signaling molecule. Anoxic environments routinely accumulate biologically produced ethylene at significant levels, but the organisms and mechanisms responsible have been slow to emerge. This study provides one possible route.
Abstract
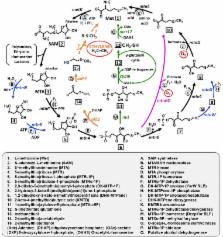
Numerous cellular processes involving S-adenosyl- l-methionine result in the formation of the toxic by-product, 5′-methylthioadenosine (MTA). To prevent inhibitory MTA accumulation and retain biologically available sulfur, most organisms possess the “universal” methionine salvage pathway (MSP). However, the universal MSP is inherently aerobic due to a requirement of molecular oxygen for one of the key enzymes. Here, we report the presence of an exclusively anaerobic MSP that couples MTA metabolism to ethylene formation in the phototrophic bacteria Rhodospirillum rubrum and Rhodopseudomonas palustris. In vivo metabolite analysis of gene deletion strains demonstrated that this anaerobic MSP functions via sequential action of MTA phosphorylase (MtnP), 5-(methylthio)ribose-1-phosphate isomerase (MtnA), and an annotated class II aldolase-like protein (Ald2) to form 2-(methylthio)acetaldehyde as an intermediate. 2-(Methylthio)acetaldehyde is reduced to 2-(methylthio)ethanol, which is further metabolized as a usable organic sulfur source, generating stoichiometric amounts of ethylene in the process. Ethylene induction experiments using 2-(methylthio)ethanol versus sulfate as sulfur sources further indicate anaerobic ethylene production from 2-(methylthio)ethanol requires protein synthesis and that this process is regulated. Finally, phylogenetic analysis reveals that the genes corresponding to these enzymes, and presumably the pathway, are widespread among anaerobic and facultatively anaerobic bacteria from soil and freshwater environments. These results not only establish the existence of a functional, exclusively anaerobic MSP, but they also suggest a possible route by which ethylene is produced by microbes in anoxic environments.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents
- Record: found
- Abstract: found
- Article: not found
Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris.
- Record: found
- Abstract: found
- Article: not found
