- Record: found
- Abstract: found
- Article: found
Norovirus Contamination Levels in Ground Water Treatment Systems Used for Food-Catering Facilities in South Korea
Read this article at
Abstract
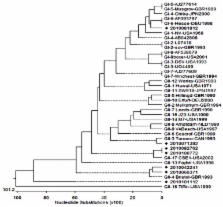
This study aimed to inspect norovirus contamination of groundwater treatment systems used in food-catering facilities located in South Korea. A nationwide study was performed in 2010. Water samples were collected and, for the analysis of water quality, the temperature, pH, turbidity, and residual chlorine content were assessed. To detect norovirus genotypes GI and GII, RT-PCR and semi-nested PCR were performed with specific NV-GI and NV-GII primer sets, respectively. The PCR products amplified from the detected strains were then subjected to sequence analyses. Of 1,090 samples collected in 2010, seven (0.64%) were found to be norovirus-positive. Specifically, one norovirus strain was identified to have the GI-6 genotype, and six GII strains had the GII, GII-3, GII-4, and GII-17 genotypes. The very low detection rate of norovirus most likely reflects the preventative measures used. However, this virus can spread rapidly from person to person in crowded, enclosed places such as the schools investigated in this study. To promote better public health and sanitary conditions, it is necessary to periodically monitor noroviruses that frequently cause epidemic food poisoning in South Korea.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Viral Gastroenteritis Outbreaks in Europe, 1995–2000
- Record: found
- Abstract: found
- Article: not found
Detection and Molecular Characterization of a Canine Norovirus
- Record: found
- Abstract: found
- Article: not found