- Record: found
- Abstract: found
- Article: found
Effects of a Common Eight Base Pairs Duplication at the Exon 7-Intron 7 Junction on Splicing, Expression, and Function of OCT1
Read this article at
Abstract
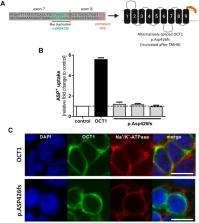
Organic cation transporter 1 (OCT1, SLC22A1) is localized in the sinusoidal membrane of human hepatocytes and mediates hepatic uptake of weakly basic or cationic drugs and endogenous compounds. Common amino acid substitutions in OCT1 were associated with altered pharmacokinetics and efficacy of drugs like sumatriptan and fenoterol. Recently, the common splice variant rs35854239 has also been suggested to affect OCT1 function. rs35854239 represents an 8 bp duplication of the donor splice site at the exon 7-intron 7 junction. Here we quantified the extent to which this duplication affects OCT1 splicing and, as a consequence, the expression and the function of OCT1. We used pyrosequencing and deep RNA-sequencing to quantify the effect of rs35854239 on splicing after minigene expression of this variant in HepG2 and Huh7 cells and directly in human liver samples. Further, we analyzed the effects of rs35854239 on OCT1 mRNA expression in total, localization and activity of the resulting OCT1 protein, and on the pharmacokinetics of sumatriptan and fenoterol. The 8 bp duplication caused alternative splicing in 38% (deep RNA-sequencing) to 52% (pyrosequencing) of the minigene transcripts when analyzed in HepG2 and Huh7 cells. The alternatively spliced transcript encodes for a truncated protein that after transient transfection in HEK293 cells was not localized in the plasma membrane and was not able to transport the OCT1 model substrate ASP +. In human liver, however, the alternatively spliced OCT1 transcript was detectable only at very low levels (0.3% in heterozygous and 0.6% in homozygous carriers of the 8 bp duplication, deep RNA-sequencing). The 8 bp duplication was associated with a significant reduction of OCT1 expression in the human liver, but explained only 9% of the general variability in OCT1 expression and was not associated with significant changes in the pharmacokinetics of sumatriptan and fenoterol. Therefore, the rs35854239 variant only partially changes splicing, causing moderate changes in OCT1 expression and may be of only limited therapeutic relevance.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found
G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences

- Record: found
- Abstract: found
- Article: found