- Record: found
- Abstract: found
- Article: found
High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems

Read this article at
Abstract
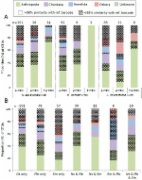
Environmental diversity surveys are crucial for the bioassessment of anthropogenic impacts on marine ecosystems. Traditional benthic monitoring relying on morphotaxonomic inventories of macrofaunal communities is expensive, time-consuming and expertise-demanding. High-throughput sequencing of environmental DNA barcodes (metabarcoding) offers an alternative to describe biological communities. However, whether the metabarcoding approach meets the quality standards of benthic monitoring remains to be tested. Here, we compared morphological and eDNA/RNA-based inventories of metazoans from samples collected at 10 stations around a fish farm in Scotland, including near-cage and distant zones. For each of 5 replicate samples per station, we sequenced the V4 region of the 18S rRNA gene using the Illumina technology. After filtering, we obtained 841,766 metazoan sequences clustered in 163 Operational Taxonomic Units (OTUs). We assigned the OTUs by combining local BLAST searches with phylogenetic analyses. We calculated two commonly used indices: the Infaunal Trophic Index and the AZTI Marine Biotic Index. We found that the molecular data faithfully reflect the morphology-based indices and provides an equivalent assessment of the impact associated with fish farms activities. We advocate that future benthic monitoring should integrate metabarcoding as a rapid and accurate tool for the evaluation of the quality of marine benthic ecosystems.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
Environmental DNA for wildlife biology and biodiversity monitoring.
- Record: found
- Abstract: found
- Article: not found