- Record: found
- Abstract: found
- Article: found
The ORF2 protein of Fusarium graminearum virus 1 suppresses the transcription of FgDICER2 and FgAGO1 to limit host antiviral defences
Read this article at
Summary
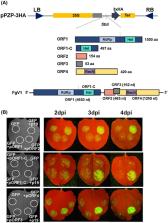
The filamentous fungus Fusarium graminearum possesses an RNA‐interference (RNAi) pathway that acts as a defence response against virus infections and exogenous double‐stranded (ds) RNA. Fusarium graminearum virus 1 (FgV1), which infects F. graminearum, confers hypovirulence‐associated traits such as reduced mycelial growth, increased pigmentation and reduced pathogenicity. In this study, we found that FgV1 can suppress RNA silencing by interfering with the induction of FgDICER2 and FgAGO1, which are involved in RNAi antiviral defence and the hairpin RNA/RNAi pathway in F. graminearum. In an FgAGO1‐ or FgDICER2‐promoter/GFP‐reporter expression assay the green fluorescent protein (GFP) transcript levels were reduced in FgV1‐infected transformed mutant strains. By comparing transcription levels of FgDICER2 and FgAGO1 in fungal transformed mutants expressing each open reading frame (ORF) of FgV1 with or without a hairpin RNA construct, we determined that reduction of FgDICER2 and FgAGO1 transcript levels requires only the FgV1 ORF2‐encoded protein (pORF2). Moreover, we confirmed that the pORF2 binds to the upstream region of FgDICERs and FgAGOs in vitro. These combined results indicate that the pORF2 of FgV1 counteracts the RNAi defence response of F. graminearum by interfering with the induction of FgDICER2 and FgAGO1 in a promoter‐dependent manner.
Abstract
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions.
- Record: found
- Abstract: found
- Article: not found
viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence.
- Record: found
- Abstract: found
- Article: not found
