- Record: found
- Abstract: found
- Article: found
Versatile protein tagging in cells with split fluorescent protein

Read this article at
Abstract
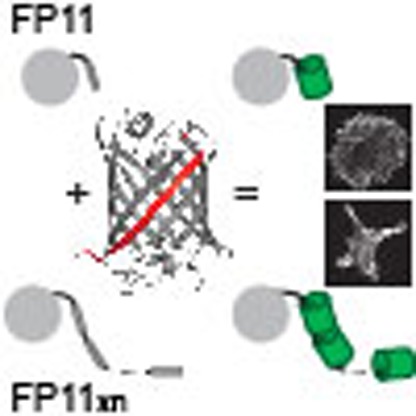
In addition to the popular method of fluorescent protein fusion, live cell protein imaging has now seen more and more application of epitope tags. The small size of these tags may reduce functional perturbation and enable signal amplification. To address their background issue, we adapt self-complementing split fluorescent proteins as epitope tags for live cell protein labelling. The two tags, GFP11 and sfCherry11 are derived from the eleventh β-strand of super-folder GFP and sfCherry, respectively. The small size of FP11-tags enables a cost-effective and scalable way to insert them into endogenous genomic loci via CRISPR-mediated homology-directed repair. Tandem arrangement FP11-tags allows proportional enhancement of fluorescence signal in tracking intraflagellar transport particles, or reduction of photobleaching for live microtubule imaging. Finally, we show the utility of tandem GFP11-tag in scaffolding protein oligomerization. These experiments illustrate the versatility of FP11-tag as a labelling tool as well as a multimerization-control tool for both imaging and non-imaging applications.
Abstract
 Tagging proteins with fluorescent proteins is a powerful method for both imaging and
non-imaging applications. Here the authors use the eleventh β-strand of sfGFP and
sfCherry as epitope tags for multicolour imaging and amplified signals by tandem arrangement;
shortness of the tag enabled introduction into genomic loci using CRISPR/Cas9.
Tagging proteins with fluorescent proteins is a powerful method for both imaging and
non-imaging applications. Here the authors use the eleventh β-strand of sfGFP and
sfCherry as epitope tags for multicolour imaging and amplified signals by tandem arrangement;
shortness of the tag enabled introduction into genomic loci using CRISPR/Cas9.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein.
- Record: found
- Abstract: found
- Article: not found
Fluorescent probes for super-resolution imaging in living cells.
- Record: found
- Abstract: found
- Article: not found