- Record: found
- Abstract: found
- Article: found
Improved split fluorescent proteins for endogenous protein labeling
Read this article at
Abstract
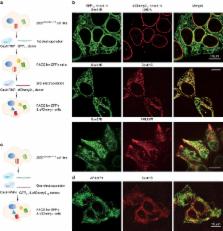
Self-complementing split fluorescent proteins (FPs) have been widely used for protein labeling, visualization of subcellular protein localization, and detection of cell–cell contact. To expand this toolset, we have developed a screening strategy for the direct engineering of self-complementing split FPs. Via this strategy, we have generated a yellow–green split-mNeonGreen2 1–10/11 that improves the ratio of complemented signal to the background of FP 1–10-expressing cells compared to the commonly used split GFP 1–10/11; as well as a 10-fold brighter red-colored split-sfCherry2 1–10/11. Based on split sfCherry2, we have engineered a photoactivatable variant that enables single-molecule localization-based super-resolution microscopy. We have demonstrated dual-color endogenous protein tagging with sfCherry2 11 and GFP 11, revealing that endoplasmic reticulum translocon complex Sec61B has reduced abundance in certain peripheral tubules. These new split FPs not only offer multiple colors for imaging interaction networks of endogenous proteins, but also hold the potential to provide orthogonal handles for biochemical isolation of native protein complexes.
Abstract
Split fluorescent proteins (FPs) have been widely used to visualise proteins in cells. Here the authors develop a screen for engineering new split FPs, and report a yellow-green split-mNeonGreen2 with reduced background, a red split-sfCherry2 for multicolour labeling, and its photoactivatable variant for super-resolution use.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy.
- Record: found
- Abstract: found
- Article: not found
A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum
- Record: found
- Abstract: found
- Article: not found