- Record: found
- Abstract: found
- Article: found
The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 α-enhancer activity
Read this article at
Abstract
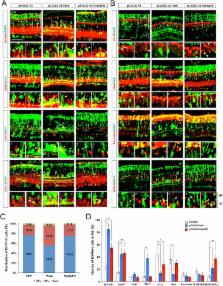
The visual responses of vertebrates are sensitive to the overall composition of retinal interneurons including amacrine cells, which tune the activity of the retinal circuitry. The expression of Paired-homeobox 6 (PAX6) is regulated by multiple cis-DNA elements including the intronic α-enhancer, which is active in GABAergic amacrine cell subsets. Here, we report that the transforming growth factor ß1-induced transcript 1 protein (Tgfb1i1) interacts with the LIM domain transcription factors Lhx3 and Isl1 to inhibit the α-enhancer in the post-natal mouse retina. Tgfb1i1 -/- mice show elevated α-enhancer activity leading to overproduction of Pax6ΔPD isoform that supports the GABAergic amacrine cell fate maintenance. Consequently, the Tgfb1i1 -/- mouse retinas show a sustained light response, which becomes more transient in mice with the auto-stimulation-defective Pax6 ΔPBS/ΔPBS mutation. Together, we show the antagonistic regulation of the α-enhancer activity by Pax6 and the LIM protein complex is necessary for the establishment of an inner retinal circuitry, which controls visual adaptation.
eLife digest
The retina is a light-sensitive layer of tissue that lines the inside of the eye. This tissue is highly organized and comprises a variety of different nerve cells, including amacrine cells. Together, these cells process incoming light and then trigger electrical signals that travel to the brain, where they are translated into an image. Changes in the nerve cell composition of the retina, or in how the cells connect to each other, can alter the visual information that travels to the brain.
The nerve cells of the retina are formed before a young animal opens its eyes for the first time. Proteins called transcription factors – which regulate the expression of genes – tightly control how the retina develops. For example, a transcription factor called Pax6 drives the development of amacrine cells. Several other transcription factors control the production of Pax6 by binding to a section of DNA known as the “α-enhancer”. However, it is not clear how regulating Pax6 production influences the development of specific sets of amacrine cells.
Kim et al. reveal that a protein known as Tgfb1i1 interacts with two transcription factors to form a “complex” that binds to the α-enhancer and blocks the production of a particular form of Pax6. In experiments performed in mice, the loss of Tgfb1i1 led to increased production of this form of Pax6, which resulted in the retina containing more of a certain type of amacrine cell that produce a molecule called GABA. Mice lacking Tgfb1i1 show a stronger response to light and are therefore comparable to people who are too sensitive to light. On the other hand, mice with a missing a section of the α-enhancer DNA have fewer amacrine cells releasing GABA and become less sensitive to light and are comparable to people who have difficulty detecting weaker light signals.
The findings of Kim et al. suggest that an individual’s sensitivity to light is related, at least in part, to the mixture of amacrine cells found in their retina, which is determined by certain transcription factors that target the α-enhancer.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Electroporation and RNA interference in the rodent retina in vivo and in vitro.
- Record: found
- Abstract: found
- Article: not found
Pax6 is required for the multipotent state of retinal progenitor cells.
- Record: found
- Abstract: found
- Article: not found