- Record: found
- Abstract: found
- Article: found
Captivity Shapes the Gut Microbiota of Andean Bears: Insights into Health Surveillance
Read this article at
Abstract
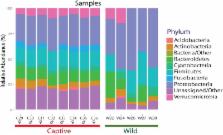
The Andean bear is an endemic species of the tropical Andes who has an almost exclusively plant-based diet. Since herbivorous mammals do not carry enzymes for fiber degradation, the establishment of symbiosis with cellulolytic microorganisms in their gastrointestinal (GI) tract is necessary to help them fulfill their nutritional needs. Furthermore, as described for other mammals, a stable, diverse, and balanced gut microbial composition is an indicator of a healthy status of the host; under disturbances this balance can be lost, leading to potential diseases of the host. The goal of this study was to describe the gut microbiota of wild and captive Andean bears and determine how habitat status influences the composition and diversity of the gut symbiotic community. Fecal samples from wild ( n = 28) and captive ( n = 8) Andean bears were collected in “Reserva Pantano de Martos” and “Fundación Bioandina”, Colombia. Composition and diversity analyses were performed using amplicons from the V4 region of the 16S rDNA gene sequenced using the Ion PGM platform. PICRUSt algorithm was applied to predict the gene content of the gut microbiome of wild and captive Andean bears. A total of 5,411 and 838 OTUs were identified for wild and captive bears, respectively. Captive bears contained a lower number of bacterial phyla ( n = 7) compared to wild individuals ( n = 9). Proteobacteria (59.03%) and Firmicutes (14.03%) were the phyla that contributed the most to differences between wild and captive bears (overall dissimilarity = 87.72%). At family level, Enterobacteriaceae drove the main differences between the two groups (13.7%). PICRUSt metagenomics predictions suggested a similar pattern of relative abundance of gene families associated with the metabolism of carbohydrates across samples in wild individuals, despite the taxonomic differences of their gut microbiota. Captivity alters the availability and diversity of food resources, which likely reduces microbiota richness and diversity compared to wild individuals. Further considerations should be taken into account for nutritional schemes improving ex-situ conservation and its potential as a surveillance tool of endangered populations of wild Andean bears.
Related collections
Most cited references36
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.
- Record: found
- Abstract: found
- Article: not found
Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences
- Record: found
- Abstract: found
- Article: not found