- Record: found
- Abstract: found
- Article: found
A novel method of microsatellite genotyping-by-sequencing using individual combinatorial barcoding
Read this article at
Abstract
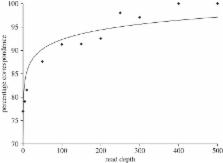
This study examines the potential of next-generation sequencing based ‘genotyping-by-sequencing’ (GBS) of microsatellite loci for rapid and cost-effective genotyping in large-scale population genetic studies. The recovery of individual genotypes from large sequence pools was achieved by PCR-incorporated combinatorial barcoding using universal primers. Three experimental conditions were employed to explore the possibility of using this approach with existing and novel multiplex marker panels and weighted amplicon mixture. The GBS approach was validated against microsatellite data generated by capillary electrophoresis. GBS allows access to the underlying nucleotide sequences that can reveal homoplasy, even in large datasets and facilitates cross laboratory transfer. GBS of microsatellites, using individual combinatorial barcoding, is potentially faster and cheaper than current microsatellite approaches and offers better and more data.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
An integrated semiconductor device enabling non-optical genome sequencing.
- Record: found
- Abstract: found
- Article: not found
The complete genome of an individual by massively parallel DNA sequencing.
- Record: found
- Abstract: found
- Article: not found