- Record: found
- Abstract: found
- Article: not found
Magnetofluid-Integrated Multicolor Immunochip for Visual Analysis of Neutralizing Antibodies to SARS-CoV-2 Variants

Abstract
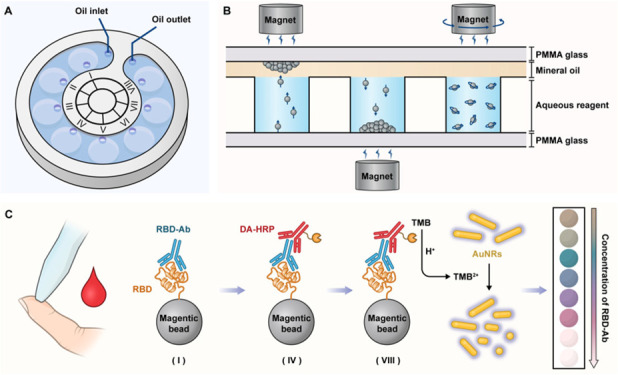
The global spread of SARS-CoV-2 virus has severely affected human health, life, and work. Vaccine immunization is considered to be an effective means to protect the body from infection. Therefore, timely analysis of the antibody level is helpful to identify people with low immune response or attenuated antibodies so as to carry out targeted and precise vaccine booster immunization. Herein, we develop a magnetofluid-integrated multicolor immunochip, as a sample-to-answer system in a fully enclosed space, for visual analysis of neutralizing antibodies of SARS-CoV-2. Generally, this chip adopts an innovative three-dimensional two-phase system that utilizes mineral oil to block the connection between reagent wells in the vertical direction and provides a wide interface for rapid and nondestructive shuttle of magnetic beads during the immunoassay. In order to obtain visualized signal output, gold nanorods with a size-dependent color effect are used as the colorful chromogenic substrates for evaluation of the antibody level. Using this chip, the neutralizing antibodies were successfully detected in vaccine-immunized volunteers with 83.3% sensitivity and 100% specificity. Furthermore, changes in antibody levels of the same individual over time were also reflected by the multicolor assay. Overall, benefiting from simple operation, airtight safety, and nonrequirement of external equipment, this platform can provide a new point-of-care testing strategy for alleviating the shortage of medical resources and promoting epidemic control in underdeveloped areas.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Human neutralizing antibodies elicited by SARS-CoV-2 infection
- Record: found
- Abstract: found
- Article: not found
Mechanisms of SARS-CoV-2 entry into cells
- Record: found
- Abstract: found
- Article: not found