- Record: found
- Abstract: found
- Article: not found
Efficacy of MEK Inhibition in Patients with Histiocytic Neoplasms
Abstract
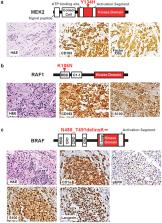
Histiocytic neoplasms are a heterogeneous group of clonal hematopoietic disorders marked by diverse mutations in the mitogen-activated protein kinase (MAPK) pathway. 1, 2 For the 50% of histiocytosis patients with BRAFV600-mutations 3– 5 , RAF inhibition is highly efficacious and has dramatically altered the natural history of the disease. 6, 7 Conversely, no standard therapy exists for the remaining 50% of patients lacking BRAFV600-mutations. While ERK dependence has been hypothesized to be a consistent feature across histiocytic neoplasms, this remains clinically unproven and many kinase mutations found in these patients have not been biologically characterized. We set out to evaluate ERK dependence in histiocytoses through a proof-of-concept clinical trial of the oral MEK1/2 inhibitor cobimetinib in patients with histiocytoses. Patients were enrolled regardless of tumor genotype. In parallel, novel MAPK alterations identified in treated patients were characterized for their ability to activate ERK. In 18 treated patients, the overall response rate (ORR) was 89% (90% CI: 73–100). Responses were durable, with no acquired resistance to date. At one year, 100% of responses were ongoing, and 94% of patients remained progression-free. Efficacy was observed regardless of genotype with responses achieved in patients with ARAF, BRAF, RAF1, NRAS, KRAS, MEK1, and MEK2 mutations. Consistent with observed responses, characterization of the novel mutations identified in treated patients confirmed them to be activating. Collectively, these data demonstrate that histiocytic neoplasms are characterized by remarkable dependence on MAPK signaling and, consequently, responsiveness to MEK inhibition. These results extend the benefits of molecularly targeted therapy to the entire spectrum of patients with histiocytosis.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Combined vemurafenib and cobimetinib in BRAF-mutated melanoma.
- Record: found
- Abstract: found
- Article: not found
Anchored multiplex PCR for targeted next-generation sequencing.
- Record: found
- Abstract: found
- Article: not found
