- Record: found
- Abstract: found
- Article: found
Web-based display of protein surface and pH-dependent properties for assessing the developability of biotherapeutics
Read this article at
Abstract
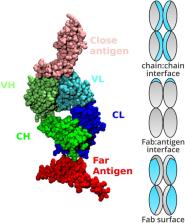
Protein instability leads to reversible self-association and irreversible aggregation which is a major concern for developing new biopharmaceutical leads. Protein solution behaviour is dictated by the physicochemical properties of the protein and the solution. Optimising protein solutions through experimental screens and targeted protein engineering can be a difficult and time consuming process. Here, we describe development of the protein-sol web server, which was previously restricted to protein solubility prediction from amino acid sequence. Tools are presented for calculating and mapping patches of hydrophobicity and charge on the protein surface. In addition, predictions of folded state stability and net charge are displayed as a heatmap for a range of pH and ionic strength conditions. Tools are evaluated in the context of antibodies, their fragments and interactions. Surprisingly, antibody-antigen interfaces are, on average, at least as polar as Fab surfaces. This benchmarking process provides the user with thresholds with which to assess non-polar surface patches, and possible solubility implications, in proteins of interest. Stability heatmaps compare favourably with experimental data for CH2 and CH3 domains. Display and quantification of surface polarity and pH/ionic strength dependence will be useful generally for investigation of protein biophysics.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies.
- Record: found
- Abstract: found
- Article: not found
Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.

- Record: found
- Abstract: found
- Article: found