- Record: found
- Abstract: found
- Article: found
Pattern of Tick Aggregation on Mice: Larger Than Expected Distribution Tail Enhances the Spread of Tick-Borne Pathogens
Read this article at
Abstract
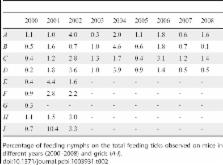
The spread of tick-borne pathogens represents an important threat to human and animal health in many parts of Eurasia. Here, we analysed a 9-year time series of Ixodes ricinus ticks feeding on Apodemus flavicollis mice (main reservoir-competent host for tick-borne encephalitis, TBE) sampled in Trentino (Northern Italy). The tail of the distribution of the number of ticks per host was fitted by three theoretical distributions: Negative Binomial (NB), Poisson-LogNormal (PoiLN), and Power-Law (PL). The fit with theoretical distributions indicated that the tail of the tick infestation pattern on mice is better described by the PL distribution. Moreover, we found that the tail of the distribution significantly changes with seasonal variations in host abundance. In order to investigate the effect of different tails of tick distribution on the invasion of a non-systemically transmitted pathogen, we simulated the transmission of a TBE-like virus between susceptible and infective ticks using a stochastic model. Model simulations indicated different outcomes of disease spreading when considering different distribution laws of ticks among hosts. Specifically, we found that the epidemic threshold and the prevalence equilibria obtained in epidemiological simulations with PL distribution are a good approximation of those observed in simulations feed by the empirical distribution. Moreover, we also found that the epidemic threshold for disease invasion was lower when considering the seasonal variation of tick aggregation.
Author Summary
Our work analyses a 9-year time series of tick co-feeding patterns on Yellow-necked mice. Our data shows a strong heterogeneity, where most mice are parasitised by a small number of ticks while few host a much larger number. We describe the number of ticks per host by the commonly used Negative Binomial model, by the Poisson-LogNormal model, and we propose the Power Law model as an alternative. In our data, the last model seems to better describe the strong heterogeneity. In order to understand the epidemiological consequences, we use a computational model to reproduce a peculiar way of transmission, observed in some cases in nature, where uninfected ticks acquire an infection by feeding on a host where infected ticks are present, without any remarkable epidemiological involvement of the host itself. In particular, we are interested in determining the conditions leading to pathogen spread. We observe that the effective transmission of this infection in nature is highly dependent on the capability of the implemented model to describe the tick burden. In addition, we also consider seasonal changes in tick aggregation on mice, showing its influence on the spread of the infection.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Heterogeneities in the transmission of infectious agents: implications for the design of control programs.
- Record: found
- Abstract: found
- Article: not found