- Record: found
- Abstract: found
- Article: found
Nascent mutant Huntingtin exon 1 chains do not stall on ribosomes during translation but aggregates do recruit machinery involved in ribosome quality control and RNA
Read this article at
Abstract
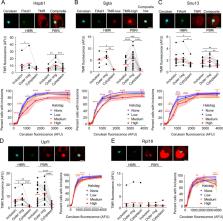
Mutations that cause Huntington’s Disease involve a polyglutamine (polyQ) sequence expansion beyond 35 repeats in exon 1 of Huntingtin. Intracellular inclusion bodies of mutant Huntingtin protein are a key feature of Huntington’s disease brain pathology. We previously showed that in cell culture the formation of inclusions involved the assembly of disordered structures of mHtt exon 1 fragments (Httex1) and they were enriched with translational machinery when first formed. We hypothesized that nascent mutant Httex1 chains co-aggregate during translation by phase separation into liquid-like disordered aggregates and then convert to more rigid, amyloid structures. Here we further examined the mechanisms of inclusion assembly in a human epithelial kidney (AD293) cell culture model. We found mHttex1 did not appear to stall translation of its own nascent chain, or at best was marginal. We also found the inclusions appeared to recruit low levels of RNA but there was no difference in enrichment between early formed and mature inclusions. Proteins involved in translation or ribosome quality control were co-recruited to the inclusions (Ltn1 Rack1) compared to a protein not anticipated to be involved (NACAD), but there was no major specificity of enrichment in the early formed inclusions compared to mature inclusions. Furthermore, we observed co-aggregation with other proteins previously identified in inclusions, including Upf1 and chaperone-like proteins Sgta and Hspb1, which also suppressed aggregation at high co-expression levels. The newly formed inclusions also contained immobile mHttex1 molecules which points to the disordered aggregates being mechanically rigid prior to amyloid formation. Collectively our findings show little evidence that inclusion assembly arises by a discrete clustering of stalled nascent chains and associated quality control machinery. Instead, the machinery appear to be recruited continuously, or secondarily, to the nucleation of inclusion formation.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo.
- Record: found
- Abstract: found
- Article: not found
Towards a transgenic model of Huntington's disease in a non-human primate.
- Record: found
- Abstract: found
- Article: not found