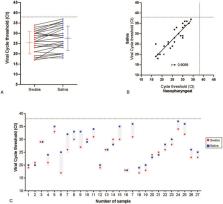
Summary Background Coronavirus disease 2019 (COVID-19) causes severe community and nosocomial outbreaks. Comprehensive data for serial respiratory viral load and serum antibody responses from patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are not yet available. Nasopharyngeal and throat swabs are usually obtained for serial viral load monitoring of respiratory infections but gathering these specimens can cause discomfort for patients and put health-care workers at risk. We aimed to ascertain the serial respiratory viral load of SARS-CoV-2 in posterior oropharyngeal (deep throat) saliva samples from patients with COVID-19, and serum antibody responses. Methods We did a cohort study at two hospitals in Hong Kong. We included patients with laboratory-confirmed COVID-19. We obtained samples of blood, urine, posterior oropharyngeal saliva, and rectal swabs. Serial viral load was ascertained by reverse transcriptase quantitative PCR (RT-qPCR). Antibody levels against the SARS-CoV-2 internal nucleoprotein (NP) and surface spike protein receptor binding domain (RBD) were measured using EIA. Whole-genome sequencing was done to identify possible mutations arising during infection. Findings Between Jan 22, 2020, and Feb 12, 2020, 30 patients were screened for inclusion, of whom 23 were included (median age 62 years [range 37–75]). The median viral load in posterior oropharyngeal saliva or other respiratory specimens at presentation was 5·2 log10 copies per mL (IQR 4·1–7·0). Salivary viral load was highest during the first week after symptom onset and subsequently declined with time (slope −0·15, 95% CI −0·19 to −0·11; R 2=0·71). In one patient, viral RNA was detected 25 days after symptom onset. Older age was correlated with higher viral load (Spearman's ρ=0·48, 95% CI 0·074–0·75; p=0·020). For 16 patients with serum samples available 14 days or longer after symptom onset, rates of seropositivity were 94% for anti-NP IgG (n=15), 88% for anti-NP IgM (n=14), 100% for anti-RBD IgG (n=16), and 94% for anti-RBD IgM (n=15). Anti-SARS-CoV-2-NP or anti-SARS-CoV-2-RBD IgG levels correlated with virus neutralisation titre (R 2>0·9). No genome mutations were detected on serial samples. Interpretation Posterior oropharyngeal saliva samples are a non-invasive specimen more acceptable to patients and health-care workers. Unlike severe acute respiratory syndrome, patients with COVID-19 had the highest viral load near presentation, which could account for the fast-spreading nature of this epidemic. This finding emphasises the importance of stringent infection control and early use of potent antiviral agents, alone or in combination, for high-risk individuals. Serological assay can complement RT-qPCR for diagnosis. Funding Richard and Carol Yu, May Tam Mak Mei Yin, The Shaw Foundation Hong Kong, Michael Tong, Marina Lee, Government Consultancy Service, and Sanming Project of Medicine.