- Record: found
- Abstract: found
- Article: found
Biological Evaluation and 3D-QSAR Studies of Curcumin Analogues as Aldehyde Dehydrogenase 1 Inhibitors
Read this article at
Abstract
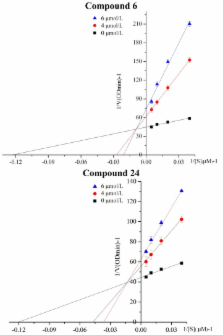
Aldehyde dehydrogenase 1 (ALDH1) is reported as a biomarker for identifying some cancer stem cells, and down-regulation or inhibition of the enzyme can be effective in anti-drug resistance and a potent therapeutic for some tumours. In this paper, the inhibitory activity, mechanism mode, molecular docking and 3D-QSAR (three-dimensional quantitative structure activity relationship) of curcumin analogues (CAs) against ALDH1 were studied. Results demonstrated that curcumin and CAs possessed potent inhibitory activity against ALDH1, and the CAs compound with ortho di-hydroxyl groups showed the most potent inhibitory activity. This study indicates that CAs may represent a new class of ALDH1 inhibitor.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Role of aldehyde dehydrogenase in cyclophosphamide-resistant L1210 leukemia.
- Record: found
- Abstract: found
- Article: not found
Alpha-glucosidase inhibition of natural curcuminoids and curcumin analogs.
- Record: found
- Abstract: found
- Article: not found