- Record: found
- Abstract: found
- Article: found
The complete chloroplast genome sequence of Helwingia himalaica (Helwingiaceae, Aquifoliales) and a chloroplast phylogenomic analysis of the Campanulidae
Read this article at
Abstract
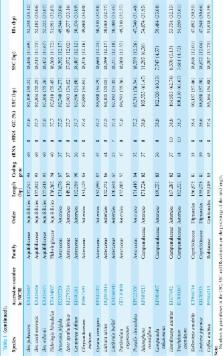
Complete chloroplast genome sequences have been very useful for understanding phylogenetic relationships in angiosperms at the family level and above, but there are currently large gaps in coverage. We report the chloroplast genome for Helwingia himalaica, the first in the distinctive family Helwingiaceae and only the second genus to be sequenced in the order Aquifoliales. We then combine this with 36 published sequences in the large (c. 35,000 species) subclass Campanulidae in order to investigate relationships at the order and family levels. The Helwingia genome consists of 158,362 bp containing a pair of inverted repeat (IR) regions of 25,996 bp separated by a large single-copy (LSC) region and a small single-copy (SSC) region which are 87,810 and 18,560 bp, respectively. There are 142 known genes, including 94 protein-coding genes, eight ribosomal RNA genes, and 40 tRNA genes. The topology of the phylogenetic relationships between Apiales, Asterales, and Dipsacales differed between analyses based on complete genome sequences and on 36 shared protein-coding genes, showing that further studies of campanulid phylogeny are needed.
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets
- Record: found
- Abstract: not found
- Article: not found
An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II
- Record: found
- Abstract: found
- Article: not found