- Record: found
- Abstract: found
- Article: not found
Cells of human aminopeptidase N (CD13) transgenic mice are infected by human coronavirus-229E in vitro, but not in vivo
Read this article at
Abstract
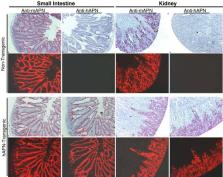
Aminopeptidase N, or CD13, is a receptor for serologically related coronaviruses of humans, pigs, and cats. A mouse line transgenic for the receptor of human coronavirus-229E (HCoV-229E) was created using human APN (hAPN) cDNA driven by a hAPN promoter. hAPN-transgenic mice expressed hAPN mRNA in the kidney, small intestine, liver, and lung. hAPN protein was specifically expressed on epithelial cells of the proximal convoluted renal tubules, bronchi, alveolar sacs, and intestinal villi. The hAPN expression pattern within transgenic mouse tissues matched that of mouse APN and was similar in mice heterozygous or homozygous for the transgene. Primary embryonic cells and bone marrow dendritic cells derived from hAPN-transgenic mice also expressed hAPN protein. Although hAPN-transgenic mice were resistant to HCoV-229E in vivo, primary embryonic cells and bone marrow dendritic cells were infected in vitro. hAPN-transgenic mice are valuable as a source of primary mouse cells expressing hAPN. This hAPN-transgenic line will also be used for crossbreeding experiments with other knockout, immune deficient, or transgenic mice to identify factors, in addition to hAPN, that are required for HCoV-229E infection.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia.
- Record: found
- Abstract: found
- Article: not found
CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus.
- Record: found
- Abstract: found
- Article: not found
