- Record: found
- Abstract: found
- Article: found
Translation elongation factor 2 depletion by siRNA in mouse liver leads to mTOR-independent translational upregulation of ribosomal protein genes
Read this article at
Abstract
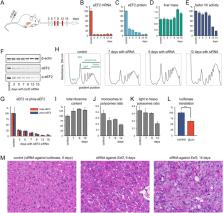
Due to breakthroughs in RNAi and genome editing methods in the past decade, it is now easier than ever to study fine details of protein synthesis in animal models. However, most of our understanding of translation comes from unicellular organisms and cultured mammalian cells. In this study, we demonstrate the feasibility of perturbing protein synthesis in a mouse liver by targeting translation elongation factor 2 (eEF2) with RNAi. We were able to achieve over 90% knockdown efficacy and maintain it for 2 weeks effectively slowing down the rate of translation elongation. As the total protein yield declined, both proteomics and ribosome profiling assays showed robust translational upregulation of ribosomal proteins relative to other proteins. Although all these genes bear the TOP regulatory motif, the branch of the mTOR pathway responsible for translation regulation was not activated. Paradoxically, coordinated translational upregulation of ribosomal proteins only occurred in the liver but not in murine cell culture. Thus, the upregulation of ribosomal transcripts likely occurred via passive mTOR-independent mechanisms. Impaired elongation sequesters ribosomes on mRNA and creates a shortage of free ribosomes. This leads to preferential translation of transcripts with high initiation rates such as ribosomal proteins. Furthermore, severe eEF2 shortage reduces the negative impact of positively charged amino acids frequent in ribosomal proteins on ribosome progression.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks.

- Record: found
- Abstract: found
- Article: found
Differential regulation of mTORC1 by leucine and glutamine
- Record: found
- Abstract: found
- Article: not found