- Record: found
- Abstract: found
- Article: found
Computational modelling of extrusion process temperatures on the interactions between black soldier fly larvae protein and corn flour starch

Read this article at
Graphical abstract
Abstract
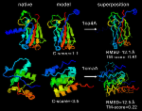
Insects such as the black soldier fly (BSF) are recently being studied as food sources to address concerns about how to meet the food demand of the growing world population, as conventional production lines for meat proteins are currently unsustainable sources. Studies have been conducted evaluating the use of insect proteins to produce extruded foods such as expanded snacks and meat analogues. However, this field of study is still quite new and not much has been studied beyond digestibility and growth performance. The purpose of this work was to evaluate the compatibility of protein extracted from BSF flour with corn flour starch within an extruded balanced shrimp feed model through molecular dynamics simulations, for which cohesive energy density and solubility parameter (δ) of both components were determined. The calculations’ results for the protein molecule systems yielded an average δ of 14.961 MPa 0.5, while the δ for starch was calculated to be 23.166 MPa 0.5. The range of difference between both δ (10 > δ > 7) suggests that the interaction of the BSF protein with corn starch is of a semi-miscible nature. These results suggest that it is possible to obtain a stable starch-protein mixture through the extrusion process.
Related collections
Most cited references51
- Record: found
- Abstract: not found
- Article: not found
The I-TASSER Suite: protein structure and function prediction.
- Record: found
- Abstract: found
- Article: not found
I-TASSER: a unified platform for automated protein structure and function prediction.
- Record: found
- Abstract: found
- Article: found
