- Record: found
- Abstract: found
- Article: found
Measuring in vivo responses to endogenous and exogenous oxidative stress using a novel haem oxygenase 1 reporter mouse
Read this article at
Abstract
Key points
-
Haem oxygenase 1 (Hmox1) is a cytoprotective enzyme with anti‐inflammatory and anti‐oxidant properties that is induced in response to multiple noxious environmental stimuli and disease states.
-
Tools to enable its expression to be monitored in vivo have been unavailable until now.
-
In a new Hmox1 reporter model we provide high‐fidelity, single‐cell resolution blueprints for Hmox1 expression throughout the body of mice.
-
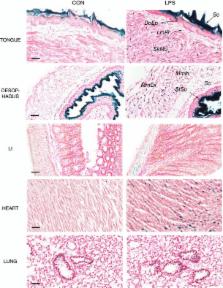
We show for the first time that Hmox1 is constitutively expressed at barrier tissues at the interface between the internal and external environments, and that it is highly induced in muscle cells during systemic inflammation.
-
These data suggest novel biological insights into the role of Hmox1 and pave the way for the use of the model to study the role of environmental stress in disease pathology.
Abstract
Hmox1 protein holds great promise as a biomarker of in vivo stress responses as it is highly induced in stressed or damaged cells. However, Hmox1 expression patterns have thus far only been available in simple model organisms with limited relevance to humans. We now report a new Hmox1 reporter line that makes it possible to obtain this information in mice, a premiere model system for studying human disease and toxicology. Using a state‐of‐the‐art strategy, we expressed multiple complementary reporter molecules from the murine Hmox1 locus, including firefly luciferase, to allow long‐term, non‐invasive imaging of Hmox1 expression, and β‐galactosidase for high‐resolution mapping of expression patterns post‐mortem. We validated the model by confirming the fidelity of reporter expression, and its responsiveness to oxidative and inflammatory stimuli. In addition to providing blueprints for Hmox1 expression in mice that provide novel biological insights, this work paves the way for the broad application of this model to establish cellular stresses induced by endogenous processes and those resulting from exposure to drugs and environmental agents. It will also enable studies on the role of oxidative stress in the pathogenesis of disease and its prevention.
Key points
-
Haem oxygenase 1 (Hmox1) is a cytoprotective enzyme with anti‐inflammatory and anti‐oxidant properties that is induced in response to multiple noxious environmental stimuli and disease states.
-
Tools to enable its expression to be monitored in vivo have been unavailable until now.
-
In a new Hmox1 reporter model we provide high‐fidelity, single‐cell resolution blueprints for Hmox1 expression throughout the body of mice.
-
We show for the first time that Hmox1 is constitutively expressed at barrier tissues at the interface between the internal and external environments, and that it is highly induced in muscle cells during systemic inflammation.
-
These data suggest novel biological insights into the role of Hmox1 and pave the way for the use of the model to study the role of environmental stress in disease pathology.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
A gene expression atlas of the central nervous system based on bacterial artificial chromosomes.
- Record: found
- Abstract: found
- Article: not found
Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise.
- Record: found
- Abstract: found
- Article: not found