- Record: found
- Abstract: found
- Article: found
Development of SNAP-Tag Fluorogenic Probes for Wash-Free Fluorescence Imaging
Read this article at
Abstract
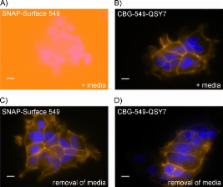
The ability to specifically attach chemical probes to individual proteins represents a powerful approach to the study and manipulation of protein function in living cells. It provides a simple, robust and versatile approach to the imaging of fusion proteins in a wide range of experimental settings. However, a potential drawback of detection using chemical probes is the fluorescence background from unreacted or nonspecifically bound probes. In this report we present the design and application of novel fluorogenic probes for labeling SNAP-tag fusion proteins in living cells. SNAP-tag is an engineered variant of the human repair protein O 6-alkylguanine-DNA alkyltransferase (hAGT) that covalently reacts with benzylguanine derivatives. Reporter groups attached to the benzyl moiety become covalently attached to the SNAP tag while the guanine acts as a leaving group. Incorporation of a quencher on the guanine group ensures that the benzylguanine probe becomes highly fluorescent only upon labeling of the SNAP-tag protein. We describe the use of intramolecularly quenched probes for wash-free labeling of cell surface-localized epidermal growth factor receptor (EGFR) fused to SNAP-tag and for direct quantification of SNAP-tagged β-tubulin in cell lysates. In addition, we have characterized a fast-labeling variant of SNAP-tag, termed SNAP f, which displays up to a tenfold increase in its reactivity towards benzylguanine substrates. The presented data demonstrate that the combination of SNAP f and the fluorogenic substrates greatly reduces the background fluorescence for labeling and imaging applications. This approach enables highly sensitive spatiotemporal investigation of protein dynamics in living cells.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
The fluorescent toolbox for assessing protein location and function.
- Record: found
- Abstract: found
- Article: not found
A general method for the covalent labeling of fusion proteins with small molecules in vivo.
- Record: found
- Abstract: found
- Article: not found