- Record: found
- Abstract: found
- Article: not found
Ectodomain shedding of TβRIII is required for TβRIII-mediated suppression of TGF-β signaling and breast cancer migration and invasion
Read this article at
Abstract
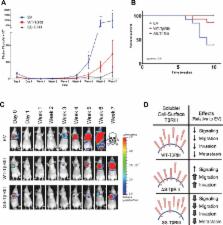
The type III TGF-β receptor (TβRIII) undergoes ectodomain shedding, with surface TβRIII enhancing and soluble TβRIII inhibiting TGF-β signaling. TβRIII mutants with impaired or enhanced shedding are used to demonstrate that the ratio of soluble to membrane-bound TβRIII regulates TβRIII/TGF-β–mediated signaling and biology in vitro and in vivo.
Abstract
The type III transforming growth factor β (TGF-β) receptor (TβRIII), also known as betaglycan, is the most abundantly expressed TGF-β receptor. TβRIII suppresses breast cancer progression by inhibiting migration, invasion, metastasis, and angiogenesis. TβRIII binds TGF-β ligands, with membrane-bound TβRIII presenting ligand to enhance TGF-β signaling. However, TβRIII can also undergo ectodomain shedding, releasing soluble TβRIII, which binds and sequesters ligand to inhibit downstream signaling. To investigate the relative contributions of soluble and membrane-bound TβRIII on TGF-β signaling and breast cancer biology, we defined TβRIII mutants with impaired (ΔShed-TβRIII) or enhanced ectodomain shedding (SS-TβRIII). Inhibiting ectodomain shedding of TβRIII increased TGF-β responsiveness and abrogated TβRIII's ability to inhibit breast cancer cell migration and invasion. Conversely, expressing SS-TβRIII, which increased soluble TβRIII production, decreased TGF-β signaling and increased TβRIII-mediated inhibition of breast cancer cell migration and invasion. Of importance, SS-TβRIII–mediated increases in soluble TβRIII production also reduced breast cancer metastasis in vivo. Taken together, these studies suggest that the ratio of soluble TβRIII to membrane-bound TβRIII is an important determinant for regulation of TβRIII- and TGF-β–mediated signaling and biology.
Related collections
Most cited references38

- Record: found
- Abstract: found
- Article: found
PROSPER: An Integrated Feature-Based Tool for Predicting Protease Substrate Cleavage Sites
- Record: found
- Abstract: found
- Article: not found
The type III TGF-beta receptor suppresses breast cancer progression.
- Record: found
- Abstract: found
- Article: not found
