- Record: found
- Abstract: found
- Article: not found
A Sunblock Based On Bioadhesive Nanoparticles
Abstract
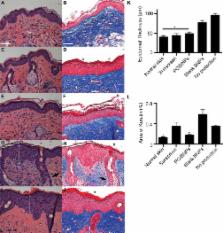
The majority of commercial sunblock preparations utilize organic or inorganic ultraviolet (UV) filters. Despite protecting against cutaneous phototoxicity, direct cellular exposure to UV filters has raised a variety of health concerns. Here, we show that the encapsulation of padimate O (PO) - a model UV filter - in bioadhesive nanoparticles (BNPs) prevents epidermal cellular exposure to UV filters while enhancing UV protection. BNPs are readily suspended in water, facilitate adherence to the stratum corneum without subsequent intra-epidermal or follicular penetration, and their interaction with skin is water-resistant yet the particles can be removed via active towel drying. Although the sunblock based on BNPs contained less than 5 wt% of the UV-filter concentration found in commercial standards, the anti-UV effect was comparable when tested in two murine models. Moreover, the BNP-based sunblock significantly reduced double-stranded DNA breaks when compared to a commercial sunscreen formulation.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies.
- Record: found
- Abstract: found
- Article: not found
Swine as models in biomedical research and toxicology testing.
- Record: found
- Abstract: found
- Article: not found
