- Record: found
- Abstract: found
- Article: found
Ex vitro hairy root induction in detached peanut leaves for plant–nematode interaction studies
Read this article at
Abstract
Background
Peanut ( Arachis hypogaea) production is largely affected by a variety of abiotic and biotic stresses, including the root-knot nematode (RKN) Meloidogyne arenaria that causes yield losses worldwide. Transcriptome studies of wild Arachis species, which harbor resistance to a number of pests and diseases, disclosed several candidate genes for M. arenaria resistance. Peanut is recalcitrant to genetic transformation, so the use of Agrobacterium rhizogenes-derived hairy roots emerged as an alternative for in-root functional characterization of these candidate genes.
Results
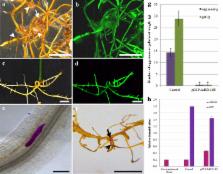
The present report describes an ex vitro methodology for hairy root induction in detached leaves based on the well-known ability of peanut to produce roots spontaneously from its petiole, which can be maintained for extended periods under high-humidity conditions. Thirty days after infection with the A. rhizogenes ‘K599’ strain, 90% of the detached leaves developed transgenic hairy roots with 5 cm of length in average, which were then inoculated with M. arenaria. For improved results, plant transformation, and nematode inoculation parameters were adjusted, such as bacterial cell density and growth stage; moist chamber conditions and nematode inoculum concentration. Using this methodology, a candidate gene for nematode resistance, AdEXLB8, was successfully overexpressed in hairy roots of the nematode-susceptible peanut cultivar ‘Runner’, resulting in 98% reduction in the number of galls and egg masses compared to the control, 60 days after M. arenaria infection.
Conclusions
This methodology proved to be more practical and cost-effective for functional validation of peanut candidate genes than in vitro and composite plant approaches, as it requires less space, reduces analysis costs and displays high transformation efficiency. The reduction in the number of RKN galls and egg masses in peanut hairy roots overexpressing AdEXLB8 corroborated the use of this strategy for functional characterization of root expressing candidate genes. This approach could be applicable not only for peanut–nematode interaction studies but also to other peanut root diseases, such as those caused by fungi and bacteria, being also potentially extended to other crop species displaying similar petiole-rooting competence.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Comprehensive algorithm for quantitative real-time polymerase chain reaction.
- Record: found
- Abstract: found
- Article: not found
Q-Gene: processing quantitative real-time RT-PCR data.
- Record: found
- Abstract: found
- Article: not found