- Record: found
- Abstract: found
- Article: not found
Microbial metabolites: cause or consequence in gastrointestinal disease?

Read this article at
Abstract
Abstract
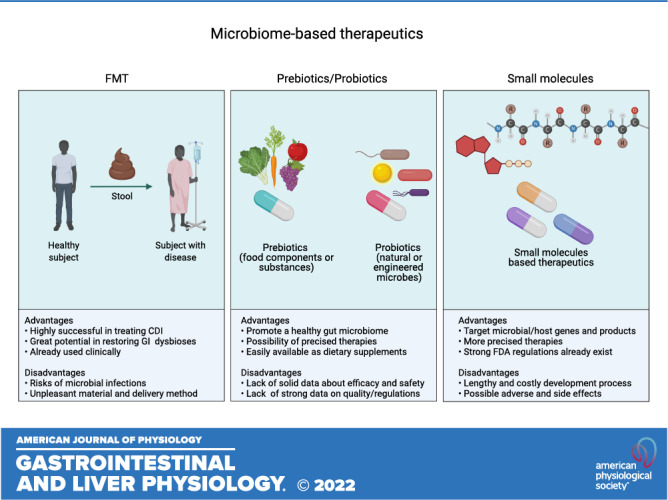
Systems biology studies have established that changes in gastrointestinal microbiome composition and function can adversely impact host physiology. Notable diseases synonymously associated with dysbiosis include inflammatory bowel diseases, cancer, metabolic disorders, and opportunistic and recurrent pathogen infections. However, there is a scarcity of mechanistic data that advances our understanding of taxonomic correlations with pathophysiological host-microbiome interactions. Generally, to survive a hostile gut environment, microbes are highly metabolically active and produce trans-kingdom signaling molecules to interact with competing microorganisms and the host. These specialized metabolites likely play important homeostatic roles, and identifying disease-specific taxa and their effector pathways can provide better strategies for diagnosis, treatment, and prevention, as well as the discovery of innovative therapeutics. The signaling role of microbial biotransformation products such as bile acids, short-chain fatty acids, polysaccharides, and dietary tryptophan is increasingly recognized, but little is known about the identity and function of metabolites that are synthesized by microbial biosynthetic gene clusters, including ribosomally synthesized and posttranslationally modified peptides (RiPPs), nonribosomal peptides (NRPs), polyketides (PKs), PK-NRP hybrids, and terpenes. Here we consider how bioactive natural products directly encoded by the human microbiome can contribute to the pathophysiology of gastrointestinal disease, cancer, autoimmune, antimicrobial-resistant bacterial and viral infections (including COVID-19). We also present strategies used to discover these compounds and the biological activities they exhibit, with consideration of therapeutic interventions that could emerge from understanding molecular causation in gut microbiome research.
Related collections
Most cited references171

- Record: found
- Abstract: found
- Article: found
Structure, Function and Diversity of the Healthy Human Microbiome
- Record: found
- Abstract: found
- Article: not found
A human gut microbial gene catalogue established by metagenomic sequencing.

- Record: found
- Abstract: found
- Article: found